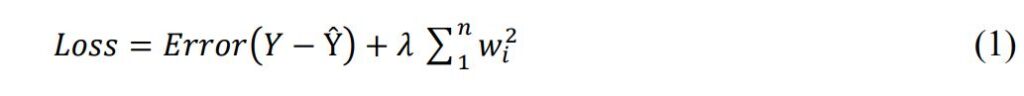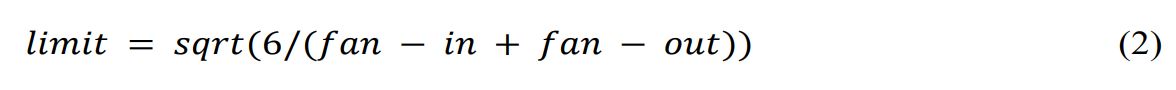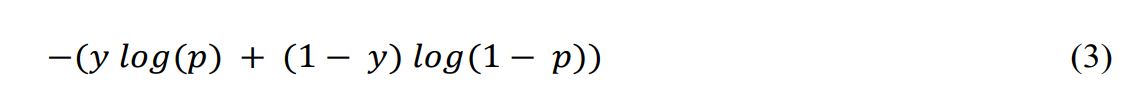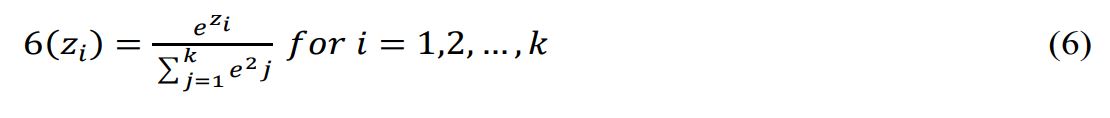The brain is an intricate organ in the human body that functions through billions of cells. Brain tumors occur due to the uncontrolled growth of cells, which can affect normal brain activities and disrupt them [1, 2]. In general, there are four main types of brain tumors, classified as grades I, II, III, and IV. Grades I and II are considered high-grade (HG) tumors, while grades III and IV are low-grade (LG) tumors. Despite significant developments in medical treatments such as surgery and chemotherapy, no effective treatment is still available for malignant brain tumors [3]. According to the reports, brain tumors are the 5th leading cause of death in women aged 20 to 39 [4]. Based on the World Health Organization (WHO), a primary brain or spinal cord tumor is a tumor that starts in the brain or spinal cord. In 2023, an estimated 24,810 adults (14,280 men and 10,530 women) in the United States will be diagnosed with primary cancerous tumors of the brain and spinal cord. Worldwide, an estimated 308,102 people were diagnosed with a primary brain or spinal cord tumor in 2020. Magnetic resonance imaging (MRI) is a non-invasive diagnostic tool used to view detailed brain structure that is the best technique to detect brain tumors [5, 6]. Brain tumors have the possibility of spreading to more parts of the body in advanced stages which are growing rapidly. Therefore, the accuracy of early diagnosis of brain tumors is vital [7, 8].
For automatic prediction, detection, and classification of brain tumors from MRI images, numerous methods and techniques have been used in recent years including discrete wavelet transform (DWT) [9], artificial neural network (ANN), and k-nearest neighbor (KNN) [10], U-Net and VGG-16 [11], LSTM-based segmentation, CapsNet, and others.
For the diagnosis of Alzheimer’s disease from MRI images, researchers proposed a pre-trained VGG-16 network model [12]. Abbasi et al. [13] presented a method in which preprocessing is performed using histogram matching and bias field correction. A convolutional neural network (CNN) model was proposed to extract features from brain MRI scans [14]. The model’s architecture included five convolutional (Conv) layers, each with a 3 × 3 filter. The model showed high accuracy for detecting pituitary tumors, but lower accuracy for meningiomas, indicating the classifier’s limited discriminative capability in this case. They achieved an overall accuracy of 81%. Totally, this work provided a fully automated classifier and does not require manually segmented tumor regions which is one of the advantages of CNN-based classifier systems. This CNN model was designed to extract features from brain MRI.
A deep CNN was utilized for monitoring brain metastases. Deep transfer learning (TL), a special deep learning (DL) class, has dominated studies on object recognition, visual categorization, and image classification problems [15]. TL enables the use of pre-trained CNN models, which were initially developed for related applications. TL has demonstrated its potential in the computer-aided diagnosis of medical problems. A pre-trained InceptionV3 model is used to differentiate between benign and malignant renal tumors in CT images [16]. Deniz et al. [17] proposed a breast cancer classifier using pre-trained VGG-16 and fine-tuned AlexNet models to extract features, followed by support vector machine (SVM) classification on histopathological images. Hussein et al. [18] presented a learning model to characterize lung and pancreatic tumors. The model architecture used for learning was a 3D CNN based on knowledge transfer. In addition, the TL-based algorithms demonstrated higher accuracy measures compared to handcrafted engineering. Hence, TL has gained significant attention in the field of neuro-oncology applications.
On brain abnormality classification, researchers used deep TL to achieve remarkable classification performance [19]. The authors trained a modified ResNet-34 model with data augmentation and fine-tuning of a TL model. Deep TL can classify medical images with minimal preprocessing, the experiment showed. The model demonstrated a method for treating brain tumor classification problems with T1-MRI images. The first significant attempt at classification utilized the challenging dataset from Figshare. The presented approach relied on manually delineated tumor borders to extract features. The authors tested various image features and classifiers. SVM model using a bag of words (BOW) features achieved superior classification performance. In this work, the performance measures used were specificity, sensitivity, and classification accuracy [20]. To solve the brain tumor classification problem, they proposed a CNN model based on DL [21].
A modified CNN architecture, known as a capsule network (CapsNet), was used in brain tumor classification by Afshar et al. [22]. In this work, the model had five learnable layers and the size of filters in all the layers was 3 × 3. The CNN model gave a classification accuracy of 81%. Using CNN features with a classifier model from the class of extreme learning machines (ELM) increased the performance. The performance improvement was meager. While CapsNet applied the spatial relationship between the tumor and its surrounding tissues.
In this work, we proposed a pre-trained CNN, the VGG-19 + TL, to classify and detect brain tumors. Also, ANN TL-based DL presented the prediction of brain tumors in four classes (meningioma, glioma, pituitary, and no-tumor) from MR images.
Dataset and pre-processing
We use the Kaggle dataset [23], which has 394 brain MRI images with one of the three tumors (meningioma, glioma, and pituitary tumors). It has 100 glioma, 115 meningioma, 105 no tumor, and 74 pituitary tumor images. Each image is (256, 256, 3). The images were pre-processed as shown in the figure (Figure 1). They were normalized to 0-1, resized to 224 × 224, converted to three channels, and cropped to remove the border.
We implemented the proposed diagnosis, classification, and prediction model in Python 3.11.3 version.

Figure 1: Data pre-processing steps.
Inductive transfer learning
Inductive TL is used when source and target tasks have the same input and output spaces, but different distributions or objectives. In this work, a model that has been trained to classify images of brain MRI can be fine-tuned to classify images of brain tumors. TL can help neural networks learn faster and better by using pre-trained models that have learned from similar or related. A model that has been trained to recognize brain tumors in images can use some of its features to recognize types of tumors like glioma, meningioma, or pituitary in new images. TL can reduce the amount of data and computation needed to train a new model, as well as improve its generalization ability. Recently, in many studies, different models based on TL have been used for the classification of images or many other cases [19, 24–26].
Convolutional neural network based-VGG-19
VGG-19 is a 19-layer deep CNN by Simonyan et al. [27]. It has 16 Conv layers and 3 fully connected layers. It uses the pre-trained VGG-19 model to extract high-level features from images for various tasks. It adds custom layers to fine-tune the base model for the specific task. For example, a dense layer with a sigmoid activation function can do binary classification. A dense layer with a Softmax activation function can do multi-class classification.
The VGG-19 model takes 224 × 224 × 3 images and has five blocks of conv and max pooling layers. Each block has two or four conv layers with more filters: 64, 128, 256, and 512. Each max pooling layer has a 2 × 2 window and a stride of 2. A flattened layer makes the output of block 5 a vector for the fully connected layer. The fully connected layer changes the input vector with weights, biases, and an activation function. It has 3 units for the three brain tumor classes: meningioma, glioma, and pituitary tumor. A Softmax layer makes the output of the fully connected layer a probability distribution over the classes. The Softmax layer gives the predicted class for the input image.
Artificial neural network
ANN is a system that mimics biological neural networks, like the human brain. It has artificial neurons that process and transmit information based on their inputs and outputs. It has multiple layers of neurons, each doing a simple math operation on its inputs. The output of each neuron is the non-linear activation function of the weighted sum of its inputs. The weights are parameters that control the connections between neurons. The input layer gets the raw data from outside. The output layer gives the final result or prediction. The hidden layers change the data through abstraction and feature extraction (Figure 2).
The ANN algorithm uses artificial neural networks to do tasks, such as classification [28], regression [29], clustering [30], and anomaly detection. It can classify brain tumor images, etc. VGG-19 can classify images well using 3 × 3 filters with the same padding, ReLU activation, and max-pooling layers.
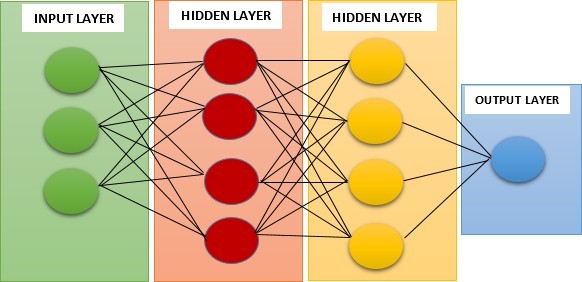
Figure 2: The structure of the ANN model.
VGG-19 + transfer learning proposed model
Brain tumor is one of the most severe diseases that affect both children and adults. It accounts for 85 to 90 percent of all the primary tumors in the Central Nervous System. Every year, about 11,700 people are diagnosed with a brain tumor. The 5-year survival rate for people with a malignant brain or CNS tumor is around 34 percent for men and 36 percent for women. Brain tumors can be classified as benign tumors, malignant tumors, pituitary tumors, etc. To improve the patient’s life expectancy, proper treatment, planning, and accurate diagnostics are essential. The best way to diagnose brain tumors is to use MRI images to train the proposed model.
The scans produce a large amount of image data, which are difficult to analyze manually by radiologists because of the complexity of brain tumors. Automated classification techniques using machine learning (ML), DL, and artificial intelligence (AI) are more precise than manual ones. Therefore, a system that uses DL algorithms, CNN, ANN, and TL for detection, classification, and prediction would assist doctors worldwide.
In this paper, we applied VGG-19 + TL to diagnose and classify brain tumors in MRI images into 2 categories and ANN + TL to predict 4 kinds of brain tumor images (glioma, meningioma, pituitary, and no-tumor). Combining VGG-19 and TL can help to classify and detect brain tumors via MRI images more accurately and efficiently.
In this paper, we apply VGG-19 + TL to classify brain tumor images from the Kaggle dataset, which has three types of brain tumors and we combine the deep features extracted by VGG-19. We also compare our approach with different ML classifiers, such as CapsNet, VGG-16, GoogLeNet, AlexNet, and Inception v3. We report that our approach achieves an accuracy of 91% for brain tumor classification and detection using the combined features. We use the ReLU activation function, which is a nonlinear function that returns the input value if it is positive, or zero otherwise. It is a widely used activation function in DL, especially for CNN. The ReLU activation function can help to avoid the vanishing gradient problem, which happens when the activation function gradients become too small and hinder the model learning. Moreover, it can induce sparsity in the network, as some neurons will not activate if their input is negative. We present our proposed model for brain tumor classification and detection in the figure below (Figure 3).
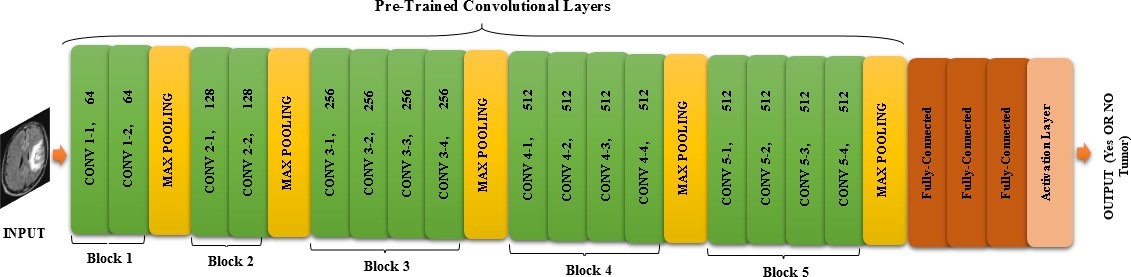
Figure 3: VGG-19 transfer learning with pre-processing and data augmentation which is proposed for the classification and detection of brain tumors via MRI images.
Our model uses VGG-19 and TL, which are two concepts related to CNN for image classification. VGG-19 is a pre-trained CNN model that can extract high-level features from any image. TL is a technique that lets us use the knowledge of a pre-trained model. VGG-19 and TL are very useful for image classification, as they can save time and resources, and achieve high accuracy. We use VGG-19 as a feature extractor, and add some custom layers to adapt it to the new task or dataset. We use our model to classify MR images of the brain into two classes (yes or no tumor) with 91.26% accuracy, as shown in the figure (Figure 4). Thresholding is an easy and effective way to segment objects of interest [31], but finding the best threshold is difficult for low-contrast images. Histogram analysis can select threshold values based on image intensity [32]. Thresholding methods can be either local or global. Global thresholding performs well when objects and backgrounds have high contrast or intensity. The optimal threshold value can be determined by the Gaussian distribution method [33]. Local thresholding is applied when a single threshold value or the entire image histogram fails to produce good segmentation results [34]. Thresholding often creates many different regions in gray-level images, as shown in the figure (Figure 5). The model layers are summarized in the table (Table 1).
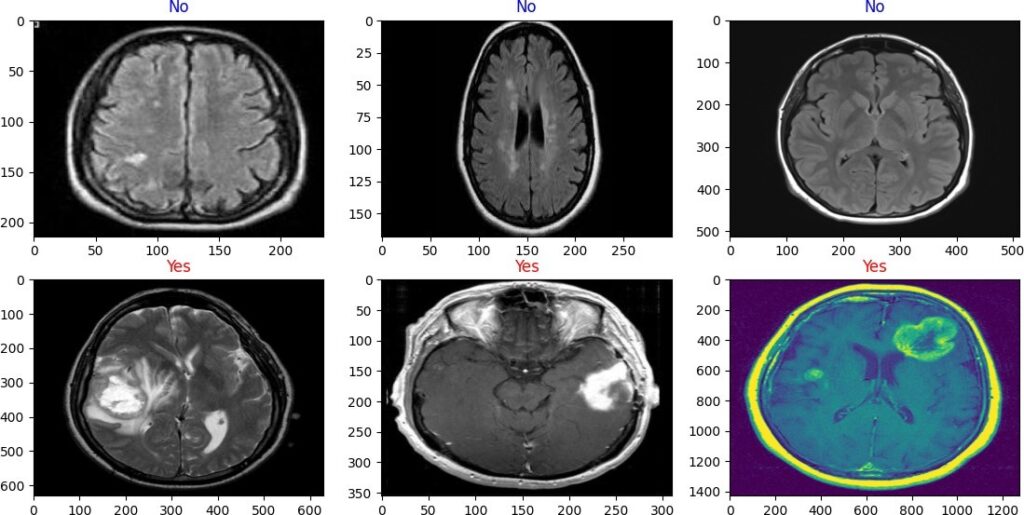
Figure 4: Classify brain tumor via 2D dimension MR images into two classes (yes and no) tumor with VGG-19 + TL model.
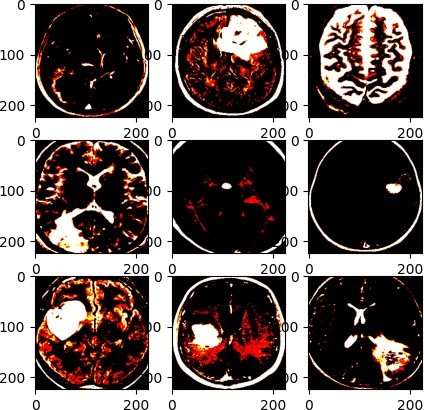
Figure 5: Thresholding of the tumors in MR images (VGG-19 + TL model).
The first layer is a pre-trained model like VGG-19 that extracts image features with an output shape of (32, 7, 7, 512). The next four layers are a conv layer with 64 3 × 3 filters, a max pooling layer that reduces the input size by half, a dropout layer that zeros 20% of the input, and a batch normalization layer that normalizes the input. The sixth layer is another conv layer with 128 3 × 3 filters and l2(0.01) regularization. The loss function of the regression model has a penalty based on the weights’ squared norm Equation 1.
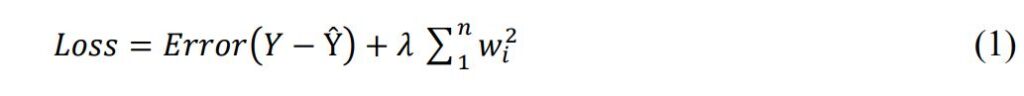
The model predicts the output or label Ŷ from the actual output or label Y. The error and regularization terms are balanced by the regularization parameter λ, which is a positive scalar that can be adjusted by cross-validation. A larger λ leads to more regularization, less overfitting, more bias, and less variance. A smaller λ leads to less regularization, more overfitting, less bias, and more variance. The model complexity is measured by the regularization term , which sums the squares of the feature weights (wi).
The next four layers are blocks of max pooling, dropout, and conv layers. The max pooling layers halve the input size by taking the max value in 2 × 2 windows. The dropout layers avoid overfitting by zeroing 20% of the input units. The conv layers use 128 filters of size 3 × 3 with ReLU activation and l2(0.01) regularization. The last layer is a flattened layer that reshapes the input into a vector for the dense layer. The dense layer has 512 units with ReLU activation and Glorot-uniform initializer.
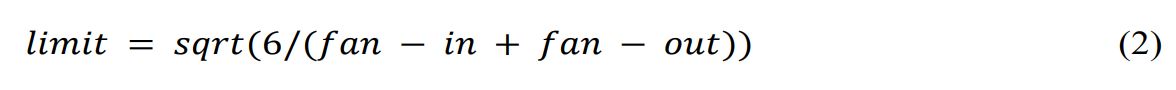
The neural network weights are initialized randomly within the range [−limit, limit], as shown in Equation 2, where limit is the variable for the bounds of the uniform distribution. The limit value is calculated by the square root function (sqrt), which depends on the input (fan − in) and output (fan − out) units in each layer. The formula includes the constant value 6.
The next seven layers are a block of dropout, batch normalization, flatten, and dense layers. The dropout layers zero 20% of the input units, the batch normalization layers normalize the input vector, the flattened layers reshape the input vector into a vector, and the dense layers have 512 units with ReLU activation and Glorot uniform initializer. The final layer is a dense layer with one unit and Softmax activation, which outputs a probability over two classes: tumor or no tumor.
In this work, we used binary classification, where the number of classes M = 2, binary cross-entropy (BCE) can be calculated as Equation 3.
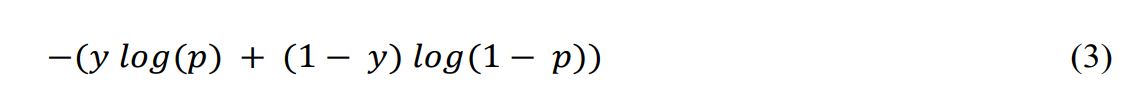
If M > 2 (i.e., multiclass classification), we calculate a separate loss for each class label per observation and sum the result Equation 4.

| Layer | Output | Param |
| VGG-19 (Functional) | (None, 7, 7, 512) | 20024384 |
| Conv2D | (None, 7, 7, 64) | 294976 |
| MaxPooling2D | (None, 7, 7, 64) | 0 |
| Dropout | (None, 7, 7, 64) | 0 |
| Batch normalization | (None, 7, 7, 64) | 256 |
| Conv2D | (None, 5, 5, 128) | 73856 |
| MaxPooling2D | (None, 5, 5, 128) | 0 |
| Dropout | (None, 5, 5, 128) | 0 |
| Conv2D | (None, 3, 3, 128) | 147584 |
| MaxPooling2D | (None, 3, 3, 128) | 0 |
| Dropout | (None, 3, 3, 128) | 0 |
| Conv2D | (None, 1, 1, 128) | 147584 |
| MaxPooling2D | (None, 1, 1, 128) | 0 |
| Dropout | (None, 1, 1, 128) | 0 |
| Flatten | (None, 128) | 0 |
| Dense | (None, 512) | 66048 |
| Dropout | (None, 512) | 0 |
| Batch normalization | (None, 512) | 2048 |
| Flatten | (None, 512) | 0 |
| Dense | (None, 512) | 262656 |
| Dropout | (None, 512) | 0 |
| Flatten | (None, 512) | 0 |
| Dense | (None, 1) | 513 |
Table 1: The summary of the proposed model (VGG-19 + TL) for classification and detection of brain tumors.
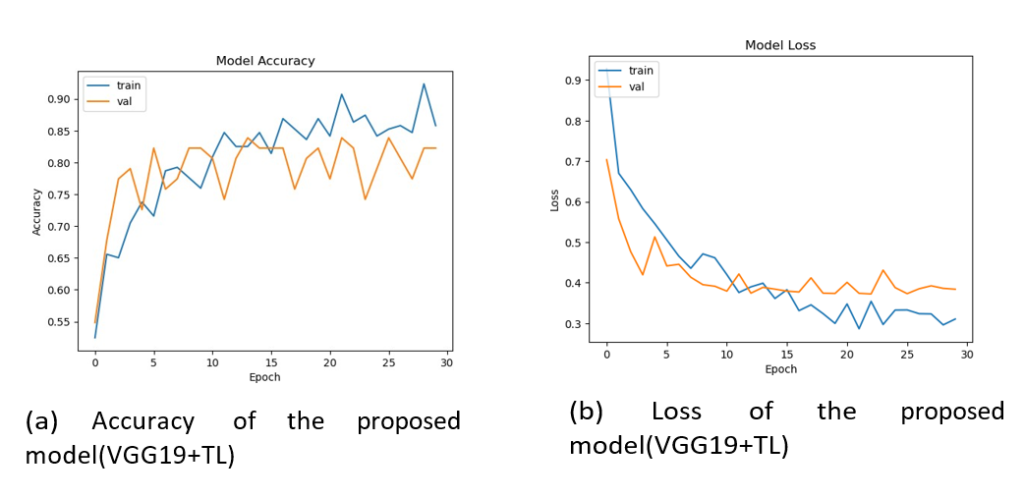
Figure 6: From left to right, (a) shows the accuracy, and (b) indicates the loss function rate of the proposed model (VGG-19 + TL).
ANN + transfer learning proposed model
Our model uses DL with TL to categorize brain tumor images into four classes. We briefly describe the model and show its predictions in the figure (Figure 7). The images undergo pre-processing steps like shuffle, crop, resize, and rotate before entering the model. We use the sequential class from TensorFlow Keras to build a linear layer stack for our model. The table (Table 2) summarizes our DL-based model. Our model’s first layer is a 3 × 3 Conv2D with 64 filters and ReLU activation, which takes a 256 × 256 × 3 image and outputs the positive values, as shown in Equation 5.

The model has a MaxPool2D layer that downsamples by taking the 2 × 2 maxima, followed by a Dropout layer with 0.2 rate to prevent overfitting. The next six layers are Conv2D-Dropout pairs with 128 filters each, enhancing the feature maps. The eighth layer flattens the output for the Dense layer with 256 units and ReLU activation.
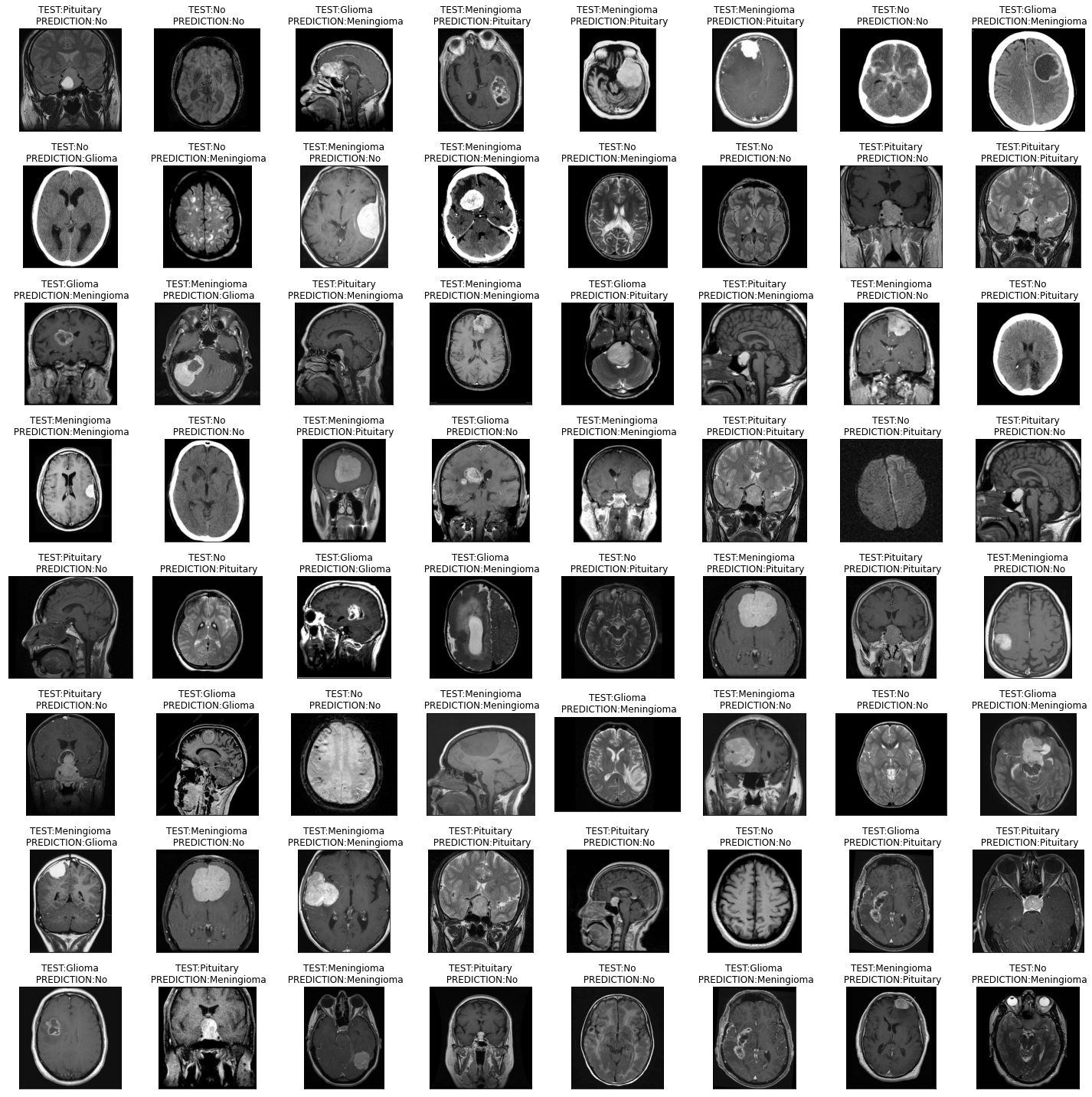
Figure 7: The output of the proposed model shows the brain MRI images which are labeled with the predicted brain tumor type.
| Layer | Output | Param |
| Input layer | (256, 256, 3) | 0 |
| Conv2d | (None, 254, 254, 64) | 1792 |
| Maxpooling2d | (None, 127, 127, 64) | 0 |
| Dropout | (None, 127, 127, 64) | 0 |
| conv2d | (None, 125, 125, 128) | 73856 |
| Maxpooling2d | (None, 62, 62, 128) | 0 |
| Dropout | (None, 62, 62, 128) | 0 |
| Conv2d | (None, 60, 60, 128) | 147584 |
| Maxpooling2d | (None, 30, 30, 128) | 0 |
| Dropout | (None, 30, 30, 128) | 0 |
| Conv2d | (None, 28, 28, 128) | 147584 |
| Maxpooling2d | (None, 14, 14, 128) | 0 |
| Dropout | (None, 14, 14, 128) | 0 |
| Flatten | (None, 25088) | 0 |
| Dense | (None, 256) | 6422784 |
| Dropout | (None, 256) | 0 |
| Dense | (None, 4) | 1028 |
Table 2: The summary of the proposed model (VGG-19 + TL) for classification and detection of brain tumors.
The last two layers are a dropout layer with 0.25 rate and a dense layer with 4 units and Softmax activation, which transforms a K-dimensional vector into probabilities, as shown in Equation 6.
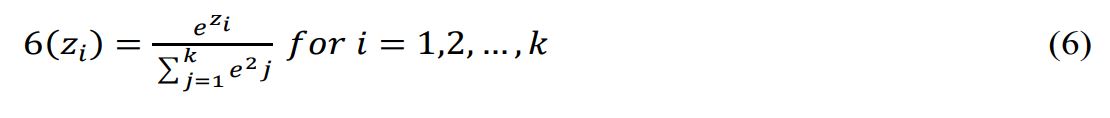
The Softmax function outputs σ(zi) for the i-th input element, which is a probability between 0 and 1 for the i-th class. It uses the exponential function ezi , which grows with zi, and the normalization term , which ensures the outputs sum to 1. zi is the score or logit of the i-th class, and K is the number of classes or inputs. This layer outputs a probability distribution over four classes. The model is compiled with RMSprop (lr = 0.001), cross-entropy loss, and accuracy metric. It is trained with the fit method and EarlyStopping callback, which stops training if the loss doesn’t improve for two epochs.
The figure (Figure 8a) shows the first experiment with ANN + TL and a pre-trained DL model on pre-processed images, achieving almost 100% accuracy on training and 91% on validation. The figure (Figure 8b) shows the accuracy and loss of the model. The figure (Figure 9 a and b) plots and summarizes the loss and accuracy on testing and validation. The figure (Figure 10 a and b) shows the confusion matrix of validation for both models. The table (Table 3) lists the parameters and settings of both models.
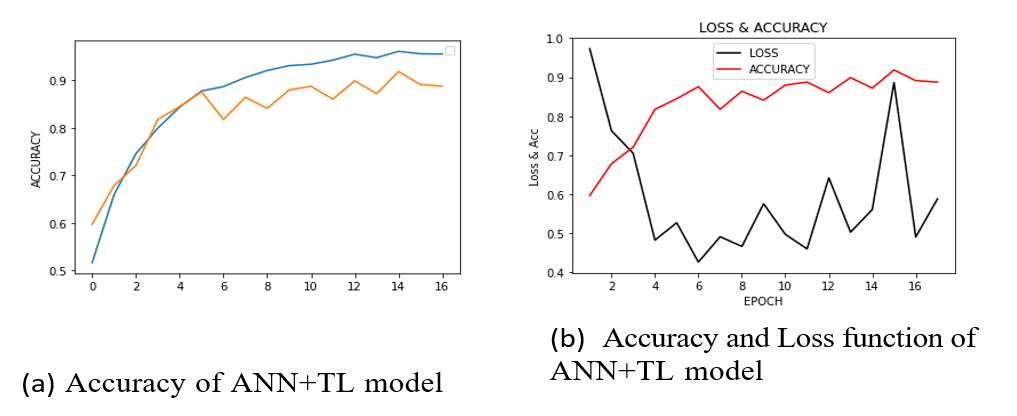
Figure 8: Both (a) and (b) show the variations of accuracy and loss function of the ANN + TL model.
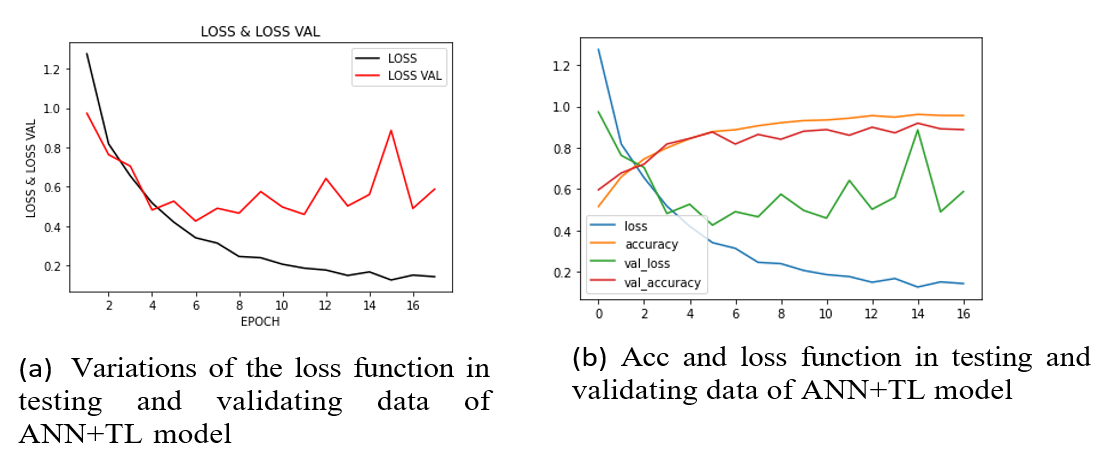
Figure 9: Both (a) and (b) illustrate the variations of the loss function in testing and validating data, and all changes of accuracy and loss function rate in a frame of ANN + TL model.
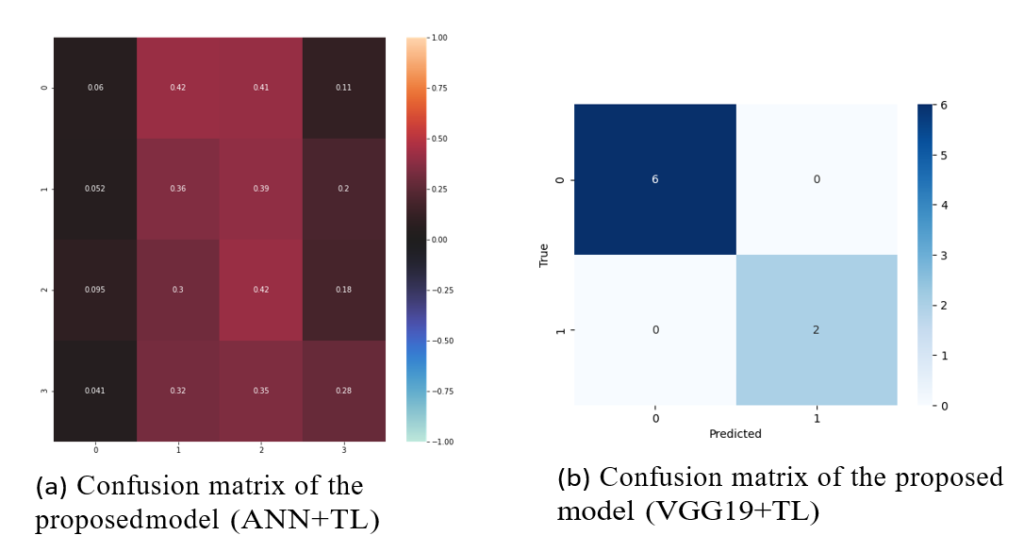
Figure 10: From left to right, (a) shows the confusion matrix of validation data related to the ANN + TL model which includes two classes (yes or no tumor), and (b) indicates the confusion matrix of validation data related to the VGG-19 + TL model which includes four classes glioma, meningioma, pituitary, and no tumor.
| Model | Parameters | Settings (values) |
| VGG-19 + TL | Initial learning rate | 0.00008 |
| Batch size | 32 |
| Epochs | 30 |
| kernel-size | (3,3) |
| kernel-regularizer | l2(0.01) |
| Bias-regularizer | l2(0.01) |
| kernel-initializer | glorot-uniform |
| Loss | binary-cross entropy |
| Optimizer | RMSprop |
| ANN + TL | Class-mode | categorical |
| Batch-size | 20 |
| Epochs | 50 |
| Seed | 42 |
| kernel-size | (3,3) |
| Activation | ReLU, Softmax |
| Input-shape | (256,256,3) |
| Pool-size | (2,2) |
| Dropout | (0.2) |
| Loss | categorical-cross entropy |
Table 3: Experimental parameters.
Performance assessment metrics
The proposed approach’s efficiency is measured by the accuracy as an evaluation metric. The classification accuracy is calculated as follows where the correctly classified high-labeled data are the true positives (TP), while the true negatives (TN) are the right predictions, although the false positives (FP) are MR images that are misclassified and the false negatives (FN) are data that are classified correctly, equation 7.

Comparison with related works
In this section, we compare our proposed method with the existing methods for diagnosing and classifying brain tumors from MRI images. We review the main features, advantages, and disadvantages of each method and highlight the differences and similarities with our method. The table (Table 4) compares related works on different datasets.
Karayegen et al. [35] proposed a DL network based on the U-Net architecture to segment the tumor region from the normal brain tissue. They reported the overall accuracy of their model for different tumor types, such as glioma, meningioma, and pituitary. However, unlike our proposed method, their method requires a large amount of annotated data for training the DL network, which is time-consuming and labor-intensive. This is a disadvantage of their method, despite the high accuracy.
Srinivas et al. [36] used a VGG-16-based CNN to classify MRI images into four classes. They achieved 92.5% accuracy, slightly higher than our VGG-19 model, but they may have overfitting issues, which can lower the model’s performance and generalization. Our models are more robust and do not overfit. We also compared our method with other techniques, such as AlexNet, U-Net, ResNet50, etc. The table (Table 4) shows the accuracy of different methods from 2019 to 2022. The challenges of this work are the limited size and diversity of the dataset, the complexity and variability of the brain tumor types, and the trade-off between the accuracy and efficiency of the models.
| Author | Method | Accuracy |
| Abiwinanda et al. [21] | CNN | 84.19% |
| Afshar et al. [22] | CapsNet | 90.89% |
| Karayegen et al. [35] | U-Net | – |
| Srinivas et al. [36] | VGG-16 | 95.5% |
| Saxena et al. [37] | VGG-16 | 90% |
| Nguyen et al. [38] | Segmentation (CNN) | 89.99% |
| C¸ inar et al. [39] | Inception v3 | 88.06% |
| GoogLeNet | 71.64% |
| AlexNet | 89.55% |
| Proposed models | VGG-19 + TL | 91.26% |
| ANN + TL | 91% |
Table 4: Related works and comparison using different datasets.
Karayegen et al. [35] propose a U-Net-based DL method for brain tumor prediction and segmentation from MR images and 3D imaging of the tumor region. They used 3064 T1-weighted images from 233 patients with three tumor types: glioma, meningioma, and pituitary. They show the accuracy and 3D visualization of their model for different tumor types. The dataset is large, but may not cover the diversity and complexity of the tumor types. The work faces challenges in the complexity and variability of the tumor types, the scarcity and diversity of the annotated data, and the accuracy-efficiency trade-off. The images are 256 × 256 × 3.
In this paper, we used a medium dataset with 2D MRI images, which has fewer parameters or features. This prevents overfitting, which can happen when the model is too sensitive to the training data. Our VGG-19 + TL model outperforms [37] in accuracy and speed, which are 90% and 500 minutes for their VGG-16 model, and 91.26% and 300 minutes for our VGG-19 + TL model. Also, our ANN + TL model runs in 100 minutes and 91% accuracy. Both our models are faster and more accurate than VGG-16.
In the table (Table 3), we show all the parameters and settings (values) that are used in both proposed models.
![]() * and Rostamy-MalKhalifeh M
* and Rostamy-MalKhalifeh M![]()